DynaBoost
Frédéric Poitevin Oliver Hoidn, PhD Alec Follmer
Selective Enhancement and Interpretation of X-ray Scattering Signals from Protein Crystals under Terahertz Excitation
Project Goal
The goal of this project is to establish a scientific program focused on studying enzyme dynamics using terahertz (THz) capabilities emerging at SLAC. It will develop a combined approach to simulating, measuring, and analyzing the scattering signal arising from THz-driven perturbations of vibrational protein motions in the crystal lattice context. Computational models will inform the design and implementation of THz-pump X-ray diffraction probe techniques at LCLS-II-HE, complementing experimental efforts underway at SSRL. Data analysis pipelines will be developed to efficiently reconstruct 3D scattering maps that will readily inform on specific structural motions at the molecular level.
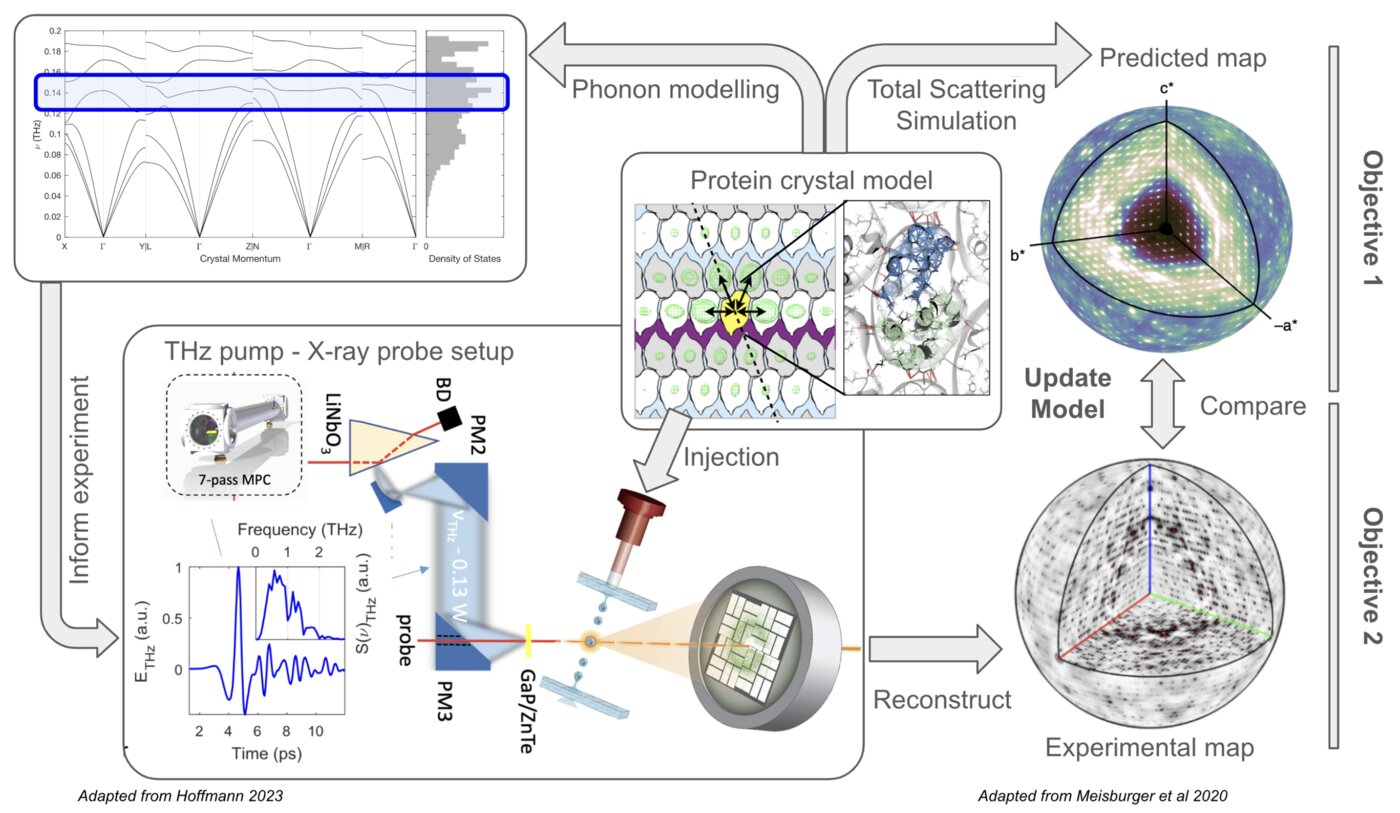
Through this proposed research, we anticipate uncovering new insights into the structural dynamics of proteins, shedding light on the fundamental processes underlying enzymatic catalysis and protein function. Specifically, exciting hypotheses in the area of protein dynamics are emerging where global collective motions of proteins and/or local electric fields within enzyme active site reaction centers may be the dominant factors that facilitate their catalytic abilities and are, in principle, at play in many protein systems. Both global collective motions and electric field effects can be studied using ultrafast excitations in the terahertz (THz) frequency range (⇠ 3 300 cm 1) providing new opportunities to expand the scope and study of enzymatic mechanisms and protein dynamics with time-resolved THz capabilities offered at SLAC. While computational and experimental support for these ideas is beginning to emerge, direct observation and quantification of these effects would represent a paradigm shift in the field of enzymology. In anticipation of these experimental capabilities at LCLS-II, this project focuses on the development and utilization of a computational framework for simulating time-resolved THz-pump X-ray diffraction experiments on protein crystals.
Recent advancements in the generation of high peak-field sources of few-cycle terahertz (THz) pulses have opened new avenues for exploring dynamic processes in solids, where the combination of single-cycle THz excitation with ultrafast X-ray diffraction probe pulses allows for the direct tracking of atomic displacements within the unit cell when driven by an intense electromagnetic field. While this type of approach has shown its potential upon application to ferroelectric crystals at LCLS, protein motions are inherently much more complex, involving non-trivial molecular re- arrangements in more porous and hydrated environments. This complexity presents a significant challenge to studying THz-induced motions that we address through a simulation-forward first-principles based approach to experimental design and data analysis.
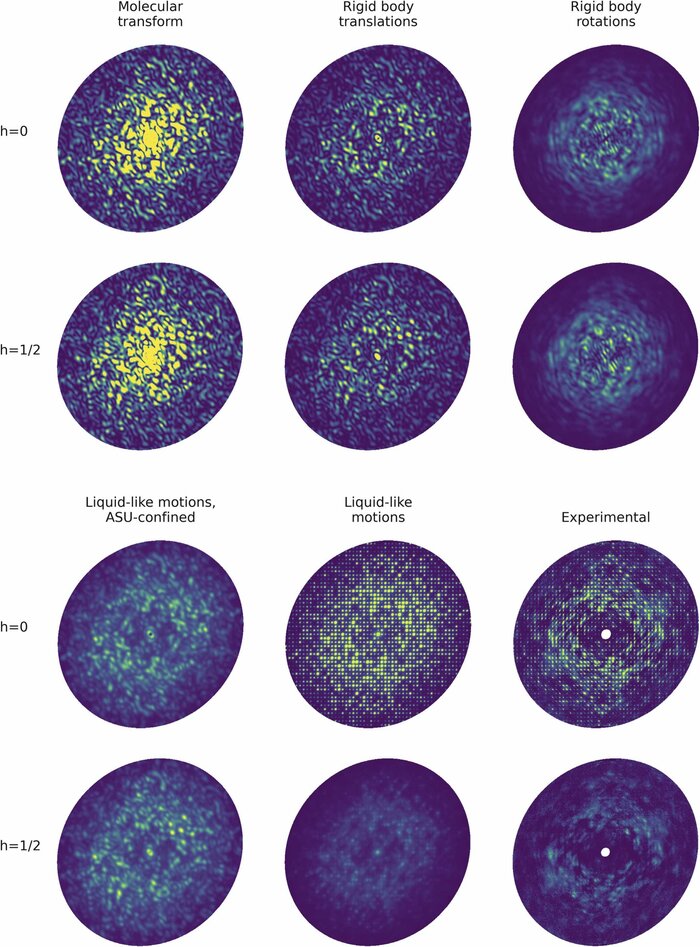
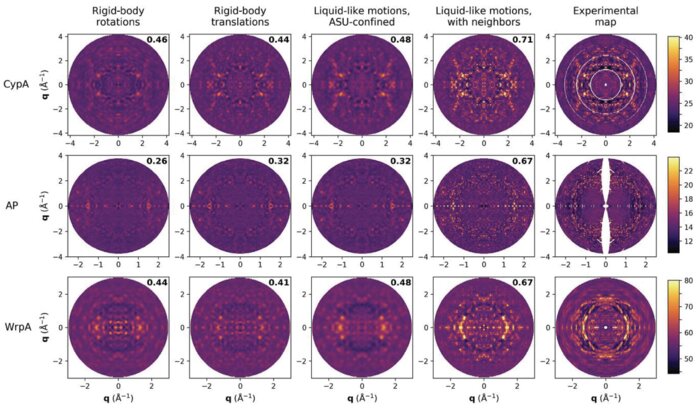
Moreover, development of a theoretical basis for these experiments will significantly aid in the interpretation and clarification of on-going diffuse X-ray scattering analyses in single-crystal protein crystallography experiments, which is a challenging and active area of investigation for studying protein dynamics (Peck et al., 2018; Peck et al., 2023). By correlating experimental observations with simulations of protein structures and their dynamics, our approach will help refine the interpretation of diffuse X-ray scattering patterns, providing enhanced clarity and precision in elucidating the structural dynamics of proteins within crystalline environments.