AtomicSPI
Frédéric Poitevin Ellen Zhong Julien Martel Gordon Wetzstein Daniel Ratner Nina Miolane Jay Shenoy Axel Levy David Klindt Ariana Peck
Learning atomic scale biomolecular dynamics from single-particle imaging data.
Project Goal
The goal of the AtomicSPI project is to deliver software helping structural biologists resolve molecular conformations from single particle imaging datasets. The project work itself consists of four connected deliverables:
- An atomic representation of the particle: A graph-based atomic model will enable the application of physics-based priors during refinement, and will be fit directly to data, rather than fitting to an intermediate reconstructed 3D map (current practice).
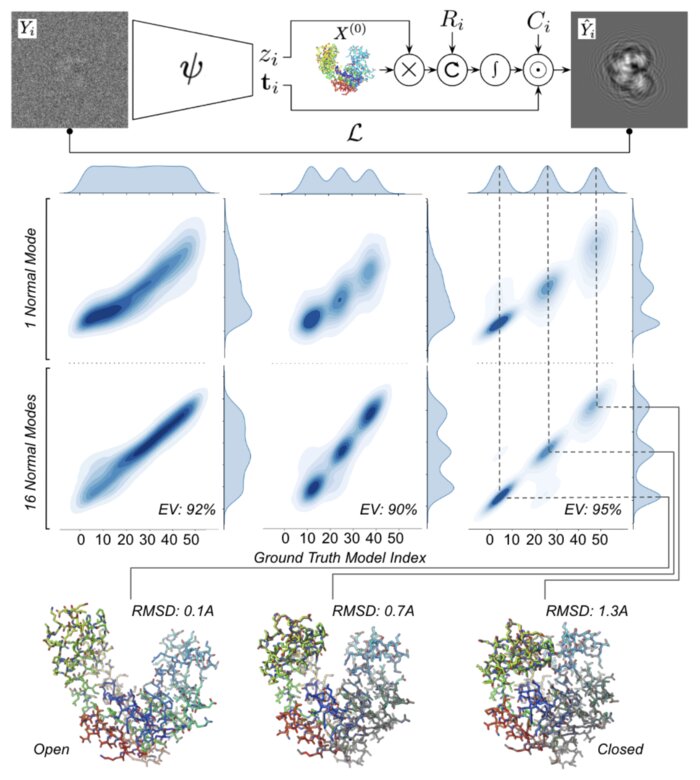
- A deep learning model that maps individual particles to a continuous space of conformations and orientations: The deep learning model will capture the molecular dynamics crucial to understanding biological function in a feasible low-dimensional space. The model will provide a distribution of conformations (i.e. the energy landscape) with applications for establishing steady-state kinetic models.
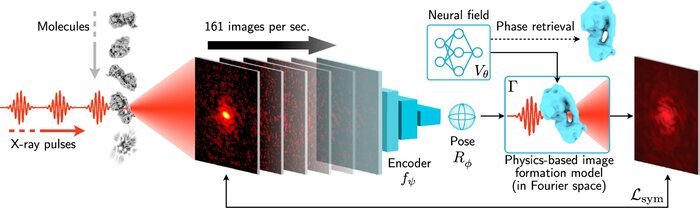
- A differentiable digital twin of the electron microscope: The simulation will map the predicted structures and orientations to the images that would be produced by the electron microscope or X-ray FEL source. Crucially, by making the simulation differentiable, it can be inverted to infer structures directly from data, proposing new structures/orientations that correspond to an experimental image.
- A deep learning reconstruction pipeline: The pipeline will tie together the above three components to learn atomic models directly from measured datasets. By combining the three components into a single step, the proposed method will be both more efficient and more accurate than existing analysis pipelines.
Accomplishments
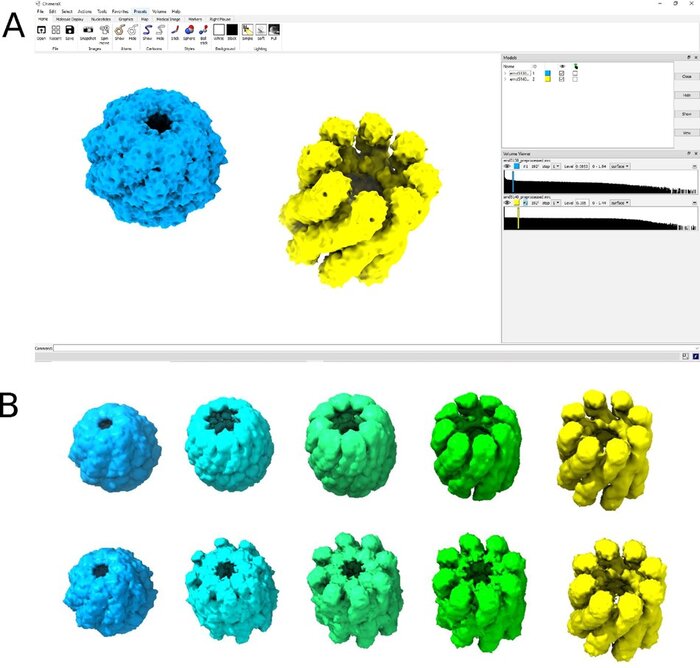
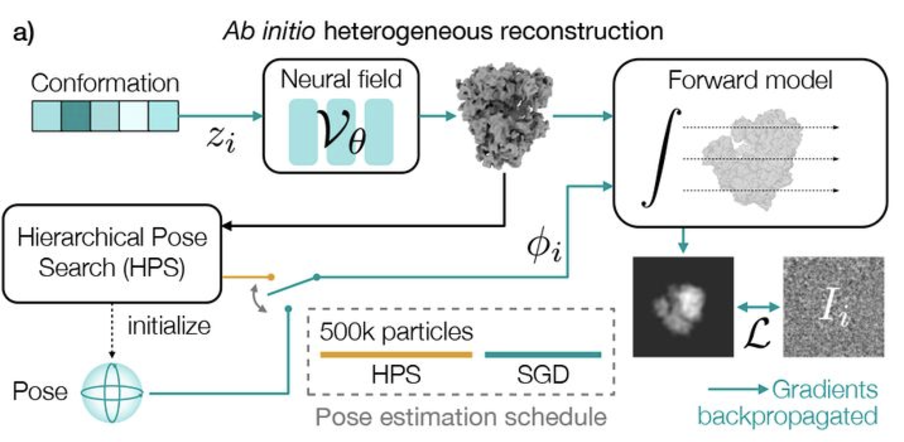
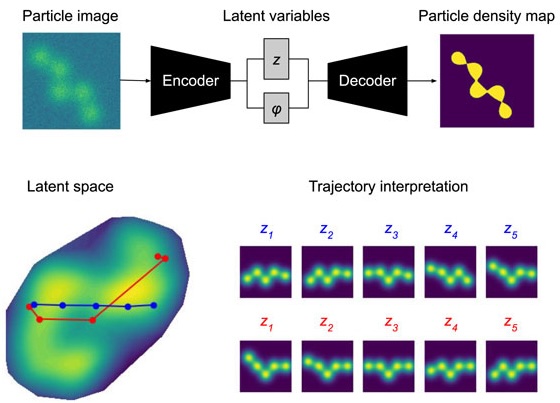
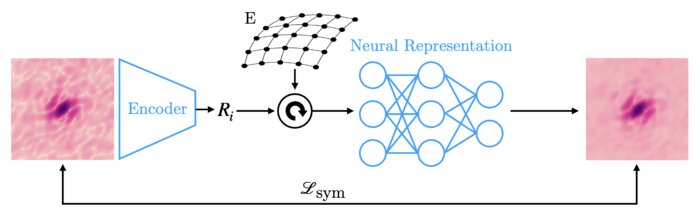
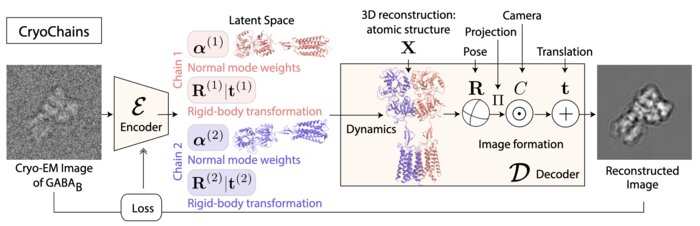
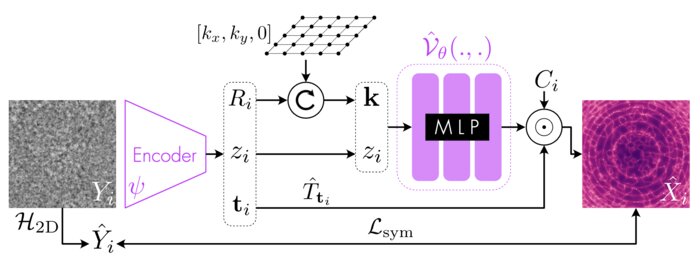
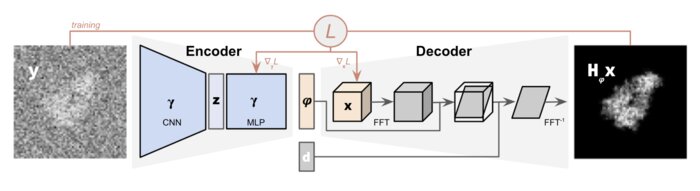
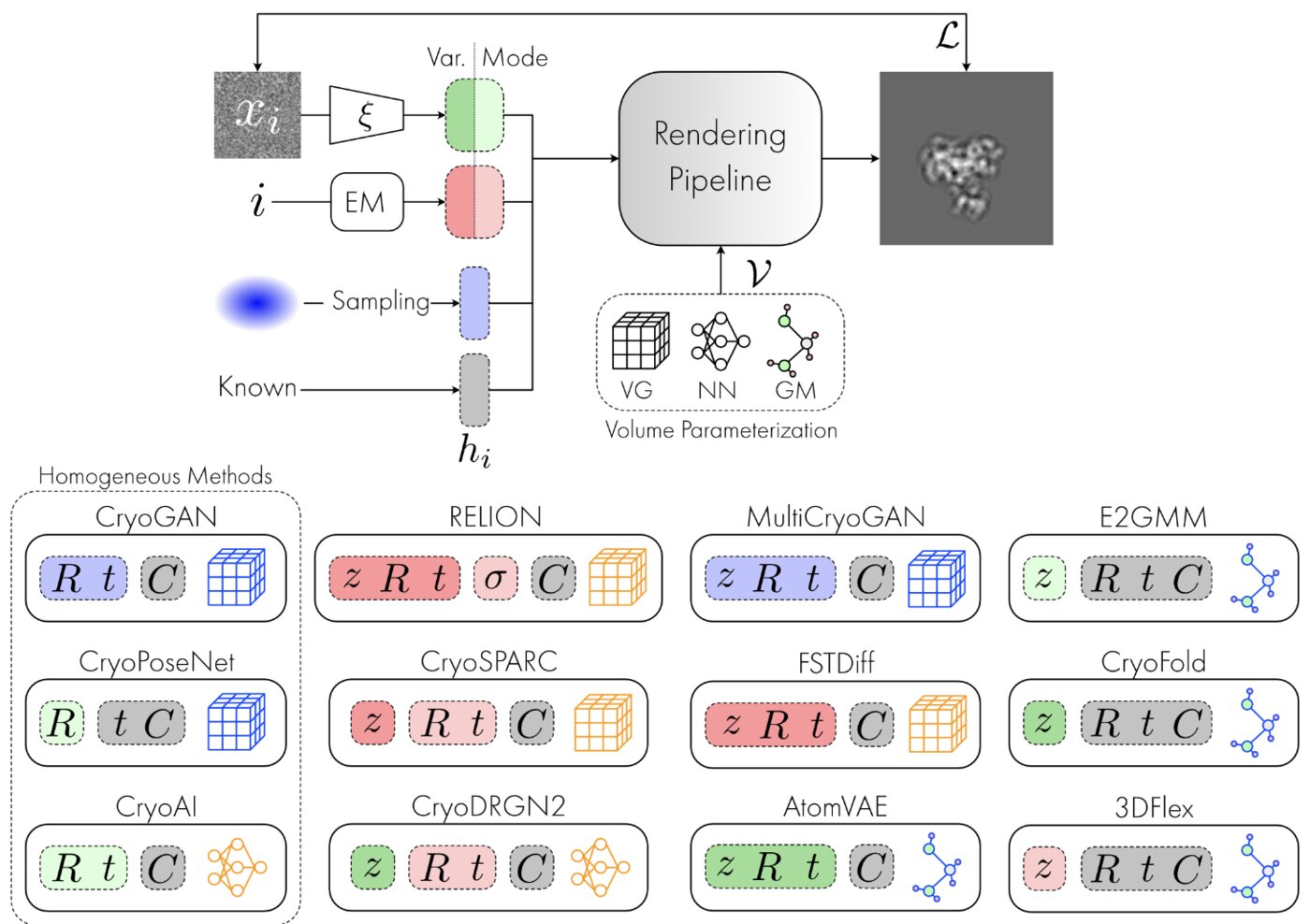
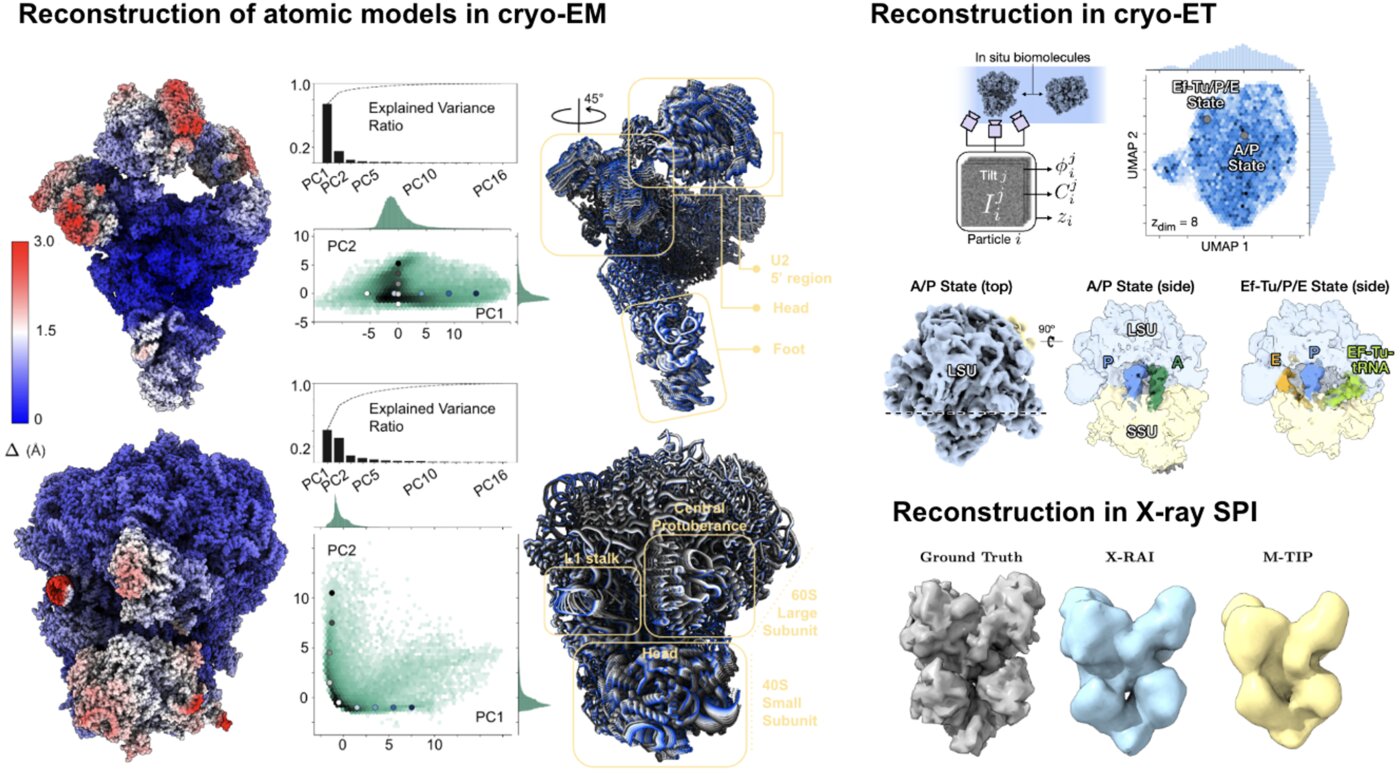
We have accomplished all four deliverables above in the cryoEM setting and prototyped them in the X-ray SPI setting. As summarized in Figure 1, the work carried thanks to this LDRD belongs to a new wave of next-generation volume reconstruction algorithm development that combines generative modeling with end-to-end unsupervised deep learning techniques (Donnat et al., 2022).
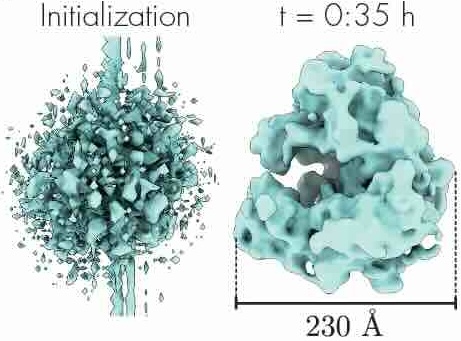
We illustrate in Figure 2 our main achievements.
AtomicSPI Projects
For a deeper dive into the AtomicSPI projects, check out their individual pages:
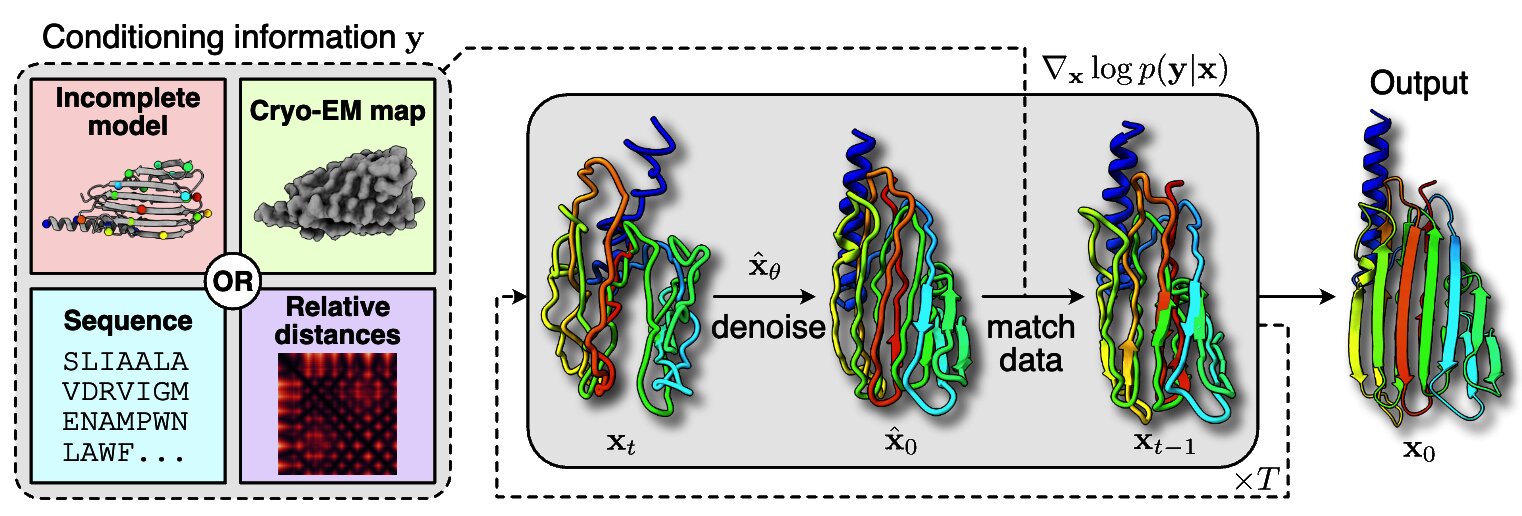
Other directions explored in the project include studies on latent disentanglement of the conformational space (Klindt et al., 2024) and a general approach to solve inverse problems in protein space using diffusion-based priors (Levy et al., 2024).
Acknowledgements
This project sprung from discussions with Nina Miolane, following our initial work described in (Miolane et al., 2020). This project was supported by the LDRD program at SLAC from 2021 to 2024.